Submitted by taire on
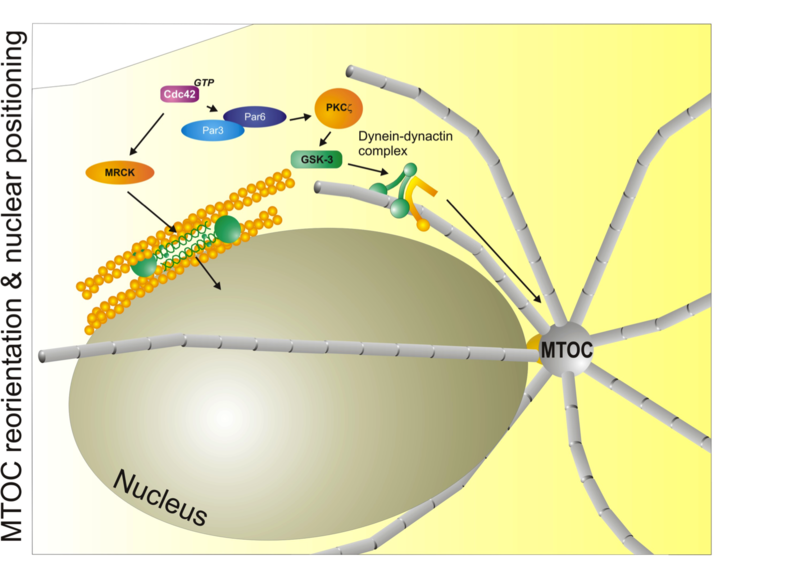 Major mechanisms that control nuclear and microtubule positioning during cell migration. The small GTPase Cdc42 acts through MRCK and myosin II to control the distance and position of the nucleus with respect to the centrosome (MTOC). Cdc42 also controls the Par3/Par6/aPKC module to control microtubule dynamics through the dynein/dynactin complex. Used with permission from Vicente-Manzanares et al., 2005 J.Cell Sci. 118,4,4917.
Major mechanisms that control nuclear and microtubule positioning during cell migration. The small GTPase Cdc42 acts through MRCK and myosin II to control the distance and position of the nucleus with respect to the centrosome (MTOC). Cdc42 also controls the Par3/Par6/aPKC module to control microtubule dynamics through the dynein/dynactin complex. Used with permission from Vicente-Manzanares et al., 2005 J.Cell Sci. 118,4,4917.In cell migration, polarity refers to the front-rear polarity - that is, the molecular and functional differences between the front (closest to the direction of migration) and rear (the opposite to the front) of the cell. Interestingly, migratory polarization shares many common effectors and adaptors with epithelial or embryonic polarity[1],[2].
Breaking symmetry
Eukaryotic cells can polarize spontaneously in the absence of pre-established asymmetric cues[3]. Polarity can also be dictated by the extracellular environment. Adhesion to the substratum or the formation of lateral contacts with other cells provides polarization cues[4]. Often, a polarized influx of soluble, extracellular signals dictates the asymmetric recruitment or activation of specific signaling proteins within the cell, creating a morphological asymmetry that translates into a functional one. For directed cell migration, gradients of soluble (chemotactic) or surface-bound/expressed (haptotactic) stimuli promote the asymmetric activation of otherwise evenly distributed cell-membrane receptors in cells that are able to migrate; this event induces polarized signals that result in the generation of a protrusion at the front of the cell (close to the highest concentration of the gradient) and a rear (close to the lowest concentration of the gradient)[5],[6]. In some cases, the composition of the plasma membrane (e.g. cholesterol-rich lipid rafts) reflects the functional polarization and acquisition of a leading and a trailing edge[7]. It is worth noting that chemoattractants often induce cellular polarization and the development of migratory, polarized morphologies even when they are present in a non-polarized, homogeneous manner. This phenomenon, called chemokinesis, reflects the intrinsic ability of migratory cells to self-polarize in response to intracellular signals or homogeneous extracellular stimuli[8].
These examples of polarization have in common the response to an extracellular stimulus that initially generates the front of the cell. However, there is strong evidence that cells can also polarize by creating the rear first[8],[9],[10]. Some cells, such as keratocytes or fibroblasts, polarize in the absence of extracellular stimulation by generating subcellular zones of 'no protrusion', regions in which protrusive signaling is inhibited and the actin cytoskeleton is reorganized into non-protrusive actin bundles[10],[11]. This mode of polarization is driven by myosin II, which has been postulated as the symmetry breaker in this model.
Forming a front
Gradients of morphogens (in the embryo), pheromones (in yeast) or chemoattractants in prospective migrating cells provide spatial cues that generate cellular asymmetry[6] by activating specific receptors. These receptors often belong to the G-protein-coupled receptor (GPCR) superfamily[12], members of which are distributed homogeneously in the cellular membrane[13],[14]. Asymmetric activation of these receptors is amplified through the asymmetric recruitment and activation of signaling adaptors in a process that magnifies very shallow differences in the gradient as perceived by the front and the rear of the cell[6].
Among the effectors that are asymmetrically recruited and activated by the membrane receptors are heterotrimeric G proteins, which activate - among other enzymes - phospholipase C (PLC) and different isoforms of protein kinase C (PKC), inducing the local formation of second messengers (such as diacylglycerol (DAG) and inositol trisphosphate (InsP3)) and protein phosphorylation[15]. G proteins also activate the phospholipid enzyme phosphoinositide 3-kinase (PI3K), which generates phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3)[16],[17]. PtdIns(3,4,5)P3 is an important second messenger in the amplification of the response to the initial gradient of stimulus and the asymmetric activation of Rho GTPases * via recruitment of guanine nucleotide-exchange factors (GEFs) that contain a pleckstrin homology (PH) domain[18]. PtdIns(3,4,5)P3 appears to be confined to the front of the cell, which is oriented towards the maximum concentration of gradient; whereas it is rapidly hydrolyzed elsewhere by the action of phosphatase and tensin homologue (PTEN), a lipid phosphatase and tumor suppressor, to maintain the gradient at the plasma membrane[19],[20].
Cdc42 is among the initial GTPases implicated in the response to polarizing signals; it controls the recruitment of PAR3 and PAR6, atypical PKCs (aPKCs) and the actin polymerization machinery to the leading edge[21]. Cdc42 participates in additional polarity-related events, such as positioning the nucleus and orienting the microtubules (see below). Genetic evidence shows that many cells can actually migrate in the absence of PI3K activity[22], whereas Cdc42 is mandatory in most systems[23],[24], suggesting that PI3K constitutes a polarity amplification mechanism[6]. It is unclear how Cdc42 is initially activated. The small Ras-related GTPase RAP1 is required for the activation of Cdc42 in some cells[25], as is the Cdc42 GEF β-PIX, in a complex with the adaptor protein Scribble[26]. Interestingly, Scribble functions downstream of the PAR3-PAR6-aPKC complex activated by Cdc42, suggesting the existence of a positive feedback loop for Cdc42 activation that probably enhances and amplifies polarization2.
Cdc42 can directly promote nucleation of dendritic networks of actin filaments via its effect on WASP and N-WASP (see below), and it also induces Rac activation through a poorly-defined mechanism[27]. The conjoined activation of Cdc42 and Rac promotes the extension of protrusions containing lamellipodia (see Protrusion section) in the direction of migration, and also regulates the additional recruitment and clustering of adhesive and chemotactic receptors and signaling molecules at the leading edge.
Bringing (up) the rear
Rac activity is inversely related to the activation of RhoA through a poorly understood feedback loop[28], such that Rho is inactive at the front of the cell while Rac mediates the formation of protrusions. RhoA activation at the rear of the cell, however, promotes the organization and contraction of actin filaments through the activation of myosin II (RLC phosphorylation on MIIAand MIIB) to form large, thick actomyosin bundles[29]. These bundles are stable (that is, they disassemble slowly) and prevent protrusion once formed, probably because they produce stable adhesions that do not signal to Rac[30]. The combination of large, stable actomyosin bundles and large adhesions results in the formation of 'no protrusion' areas at the cell rear and sides.
Cells do not always form morphologically identical rears. Differences in morphologies probably reflect different amounts of myosin II, its isoforms, and its activation state, as well as the adhesions the cells produce upon contact with the extracellular matrix[31]. For example, when a monolayer of epithelial cells is 'wounded' (by a scratch), the 'no protrusion' areas are defined by the contacts, on the sides and the rear, with the cells that surround the leading cell. Isolated fibroblasts and migrating endothelial cells display an extended rear that confers a triangular appearance on the migrating cell. In some cases, transformed variants of these cells might display disorganized rears; this phenomenon often correlates with the presence of more than one leading edge, caused by over-active protrusive signaling [32]. Keratocytes and melanocytes often display a fan-shaped morphology with a flat rear characterized by thick actomyosin bundles parallel to the leading edge[31]. Leukocytes and amoebae have short, stubby rears that accumulate small actomyosin bundles and microtubules, are often elevated over the plane of the substrate (in which case they are known as 'uropods') and accumulate adhesion receptors[33].
Other intracellular polarity systems
Other cellular organelles are also polarized in migrating cells. The nucleus, Golgi apparatus, and microtubules seem to act in concert to guide cell migration. Nuclear translocation and microtubule polarization define the direction of polarization in most mesenchymal cells but probably not in small, fast-moving cells (such as leukocytes)[34],[35]. Microtubules are polarized in part through regulation of their dynamic (growing) plus end. Microtubule capture at the cell cortex involves stabilization and capture of their plus end, and might lead to pulling forces being exerted on the microtubules[36]. Plus-end-tracking proteins (+TIPs), such as end-binding-protein-1 (EB-1), associate with the plus end of microtubules and are involved in cortical capture.
In addition to its role in controlling the chemotactic response, Cdc42 - acting through its kinase effector myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK) - positions the nucleus through myosin-dependent rearward flow of corticalactin[37]. Positioning of the microtubule organizing centre (MTOC) is regulated by Cdc42 via PAR proteins, PKCζ and its downstream effectors glycogen synthase kinase-3β (GSK-3β) and adenomatous polyposis coli (APC)[38]. These pathways regulate, among others, the dynein-dynactin cytoskeletal motor complex, and are thought to keep the MTOC immobilized with respect to the moving nucleus[37],[39]. Thus, Cdc42 is a master regulator of polarity through its role in controlling protrusion at the leading edge, nuclear positioning, and microtubule organization in response to motile cues.
- Li, R. & Gundersen, G. G. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol 9, 860-73 (2008).Pubmed abstract # 18946475
- Iden, S. & Collard, J. G. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol 9, 846-59 (2008). Pubmed abstract # 18946474
- Wedlich-Soldner, R. & Li, R. Spontaneous cell polarization: undermining determinism. 5, 267-270 (2003). Pubmed abstract # 12669070
- Parsons, J. T., Horwitz, A.R. & Schwartz, M.A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol 11, 633-43 (2010). Pubmed abstract # 20729930
- Iglesias, P. A. & Devreotes, P. N. Navigating through models of chemotaxis. Curr Opin Cell Biol 20, 35-40 (2008). Pubmed abstract # 18207721
- Swaney, K. F., Huang, C. H. & Devreotes, P. N. Eukaryotic Chemotaxis: A Network of Signaling Pathways Controls Motility, Directional Sensing, and Polarity. Annu Rev Biophys (2010). Pubmed abstract # 20192768
- Gomez-Mouton, C. et al. Dynamic redistribution of raft domains as an organizing platform for signaling during cell chemotaxis. J Cell Biol 164, 759-68 (2004). Pubmed abstract # 14981096
- Verkhovsky, A. B., Svitkina, T. M. & Borisy, G. G. Self-polarization and directional motility of cytoplasm. Curr Biol 9, 11-20 (1999). Pubmed abstract # 9889119
- Yam, P. T. et al. Actin-myosin network reorganization breaks symmetry at the cell rear to spontaneously initiate polarized cell motility. J Cell Biol 178, 1207-21 (2007). Pubmed abstract # 17893245
- Vicente-Manzanares, M., Koach, M. A., Whitmore, L., Lamers, M. L. & Horwitz, A. F. Segregation and activation of myosin IIB creates a rear in migrating cells. J Cell Biol 183, 543-54 (2008). Pubmed abstract # 18955554
- Mseka, T., Coughlin, M. & Cramer, L. P. Graded actin filament polarity is the organization of oriented actomyosin II filament bundles required for fibroblast polarization. Cell Motil Cytoskeleton 66, 743-53 (2009). Pubmed abstract # 19544402
- Cotton, M. & Claing, A. G protein-coupled receptors stimulation and the control of cell migration. Cell Signal 21, 1045-53 (2009). Pubmed abstract # 19249352
- Xiao, Z., Zhang, N., Murphy, D. B. & Devreotes, P. N. Dynamic distribution of chemoattractant receptors in living cells during chemotaxis and persistent stimulation. J Cell Biol 139, 365-74 (1997). Pubmed abstract # 9334341
- Servant, G., Weiner, O. D., Neptune, E. R., Sedat, J. W. & Bourne, H. R. Dynamics of a chemoattractant receptor in living neutrophils during chemotaxis. Mol Biol Cell 10, 1163-78 (1999). Pubmed abstract # 10198064
- Van Haastert, P. J. & Devreotes, P. N. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol 5, 626-34 (2004).Pubmed abstract # 15366706
- Kolsch, V., Charest, P. G. & Firtel, R. A. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci 121, 551-9 (2008). Pubmed abstract # 18287584
- Cain, R. J. & Ridley, A. J. Phosphoinositide 3-kinases in cell migration. Biol Cell 101, 13-29 (2009). Pubmed abstract # 19055486
- Garcia-Mata, R. & Burridge, K. Catching a GEF by its tail. Trends Cell Biol 17, 36-43 (2007). Pubmed abstract # 17126549
- Iijima, M. & Devreotes, P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109, 599-610 (2002). Pubmed abstract #12062103
- Funamoto, S., Meili, R., Lee, S., Parry, L. & Firtel, R. A. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 109, 611-23 (2002). Pubmed abstract # 12062104
- Etienne-Manneville, S. & Hall, A. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr Opin Cell Biol 15, 67-72 (2003). Pubmed abstract # 12517706
- Hoeller, O. & Kay, R. R. Chemotaxis in the absence of PIP3 gradients. Curr Biol 17, 813-7 (2007). Pubmed abstract # 17462897
- Heasman, S. J. & Ridley, A. J. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 9, 690-701 (2008). Pubmed abstract # 18719708
- Etienne-Manneville, S. Cdc42--the centre of polarity. J Cell Sci 117, 1291-300 (2004). Pubmed abstract # 15020669
- Gerard, A., Mertens, A. E., van der Kammen, R. A. & Collard, J. G. The Par polarity complex regulates Rap1- and chemokine-induced T cell polarization. J Cell Biol 176, 863-75 (2007). Pubmed abstract # 17353362
- Osmani, N., Vitale, N., Borg, J. P. & Etienne-Manneville, S. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr Biol 16, 2395-405 (2006). Pubmed abstract # 17081755
- Nobes, C. D. & Hall, A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53-62 (1995). Pubmed abstract # 7536630
- Sander, E. E., ten Klooster, J. P., van Delft, S., van der Kammen, R. A. & Collard, J. G. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol 147, 1009-22 (1999). Pubmed abstract # 10579721
- Vicente-Manzanares, M., Ma, X., Adelstein, R. S. & Horwitz, A. R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 10, 778-90 (2009). Pubmed abstract # 19851336
- Vicente-Manzanares, M., Choi, C. K. & Horwitz, A. R. Integrins in cell migration--the actin connection. J Cell Sci 122, 199-206 (2009). Pubmed abstract # 19118212
- Keren, K. et al. Mechanism of shape determination in motile cells. Nature 453, 475-80 (2008). Pubmed abstract # 18497816
- Kollega, J. The role of myosin II motor activity in distributing myosin asymmetrically and coupling protrusive activity to cell translocation. Mol Biol Cell 17, 4435-45 (2006). Pubmed abstract # 16855019
- Sanchez-Madrid, F. & Serrador, J. M. Bringing up the rear: defining the roles of the uropod. Nat Rev Mol Cell Biol 10, 353-9 (2009). Pubmed abstract # 19373240
- Omelchenko, T., Vasiliev, J. M., Gelfand, I. M., Feder, H. H. & Bonder, E. M. Mechanisms of polarization of the shape of fibroblasts and epitheliocytes: Separation of the roles of microtubules and Rho-dependent actin-myosin contractility. Proc Natl Acad Sci U S A 99, 10452-7 (2002). Pubmed abstract # 12149446
- Ratner, S., Sherrod, W. S. & Lichlyter, D. Microtubule retraction into the uropod and its role in T cell polarization and motility. J Immunol 159, 1063-7 (1997). Pubmed abstract # 9233597
- Lansbergen, G. & Akhmanova, A. Microtubule plus end: a hub of cellular activities. Traffic 7, 499-507 (2006). Pubmed abstract # 16643273
- Gomes, E. R., Jani, S. & Gundersen, G. G. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121, 451-63 (2005). Pubmed abstract # 15882626
- Etienne-Manneville, S. & Hall, A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature 421, 753-6 (2003). Pubmed abstract # 12610628
- Schmoranzer, J. et al. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol 19, 1065-74 (2009). Pubmed abstract # 19540120