Submitted by taire on
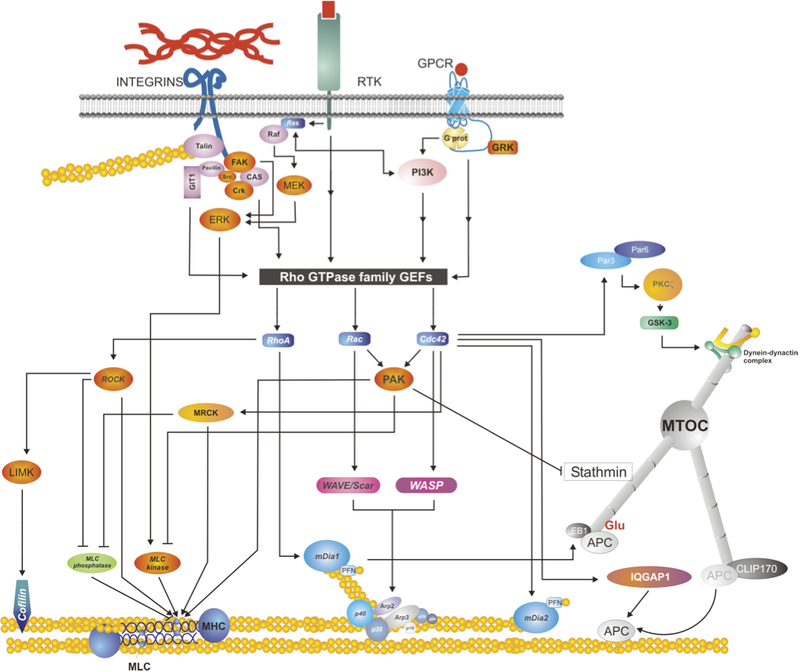
Diagram depicts several signaling pathways activated by plasma membrane receptors, including integrins (top left), receptor tyrosine kinases (e.g. EGFR, top center) and G-protein coupled receptors (top right). The diagram emphasizes a signaling convergence point, the GTPase activators/Rho GTPases (in black/blue, center of the picture), and their endpoints: from bottom left to right: cofilin, which regulates actin depolymerization; myosin II, which organizes and remodels actin; actin polymerization nucleators and promoters (Arp2/3 and Dia1/2, respectively); and molecules that interact and/or regulate microtubules (e.g. stathmin, the EBx/APC complex). Used with permission from Vicente-Manzanares et al., 2005 J.Cell Sci. 118,4,4917. click to enlarge
Cell migration requires the tight spatiotemporal regulation of multiple intertwined signaling networks that are initiated and organized at the cell surface. Signaling can be initiated through G-coupled-protein receptors (GPCRs), integrins, growth factors and other receptors (for example, cadherins, neuronal and immune receptors, and so on). The complexity of this interconnected network is seen in the synergy between growth factor receptor and adhesion receptor signaling, for example. See the figure for a simplified outline of some of the signaling intermediates between the receptors, the Rho GTPases and the migration machinery. Many of these receptors aggregate in response to the extracellular stimulus and ultimately organize signaling scaffolds. Responses at the level of the receptor include: an intrinsic kinase activity in some growth factor receptors, leading to either autophosphorylation or phosphorylation of proximal effectors. In other cases, activation occurs via conformational change upon ligand binding, which triggers binding to (or dissociation from) downstream effectors. These initial signals are then propagated along one or more branches of a complex, interconnected network of intermediates that eventually reach the Rho family of small GTPases. Small Rho (Ras-homology) GTPases are central to cell migration and act through a myriad of effectors on a relatively small number of endpoints to regulate, among other processes, the polymerization, organization and contraction of actin, polymerization and/or stability of microtubules, and transcriptional regulation of motogenic gene products (see separate box on Rho GTPases for further information).
Several guanine nucleotide-exchange factor (GEFs) can activate one or more Rho GTPases specifically[1]; similarly, many GTPase-activating proteins (GAPs) can catalyze GTP hydrolysis by one or more Rho GTPase proteins. Additionally, GDP-dissociation inhibitors (GDIs) can sequester specific Rho GTPases in the cytoplasm. In a similar fashion, specific Rho GTPases control specific effectors (kinases, actin polymerization nucleators, and so on), such that each GTPase can control different aspects of the cellular organization required for efficient cell migration.
In addition to the Rho GTPases, other small GTPases are implicated in migration. Ras activates phosphoinositide 3-kinase (PI3K), which is involved in cell migration towards an extracellular stimulus. A close analogue of Ras, Rap, controls integrin affinity (and, thereby, cell adhesion) via its effectors RapL and RIAM[2]. Members of the Rab/ARF family control vesicular trafficking and are implicated in the recycling of adhesion receptors from and to the plasma membrane during cell migration in order for the process to continue[3].
- Rossman, K. L., Der, C. J. & Sondek, J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 6, 167-80 (2005). Pubmed abstract # 15688002
- Katagiri, K., Maeda, A., Shimonaka, M. & Kinashi, T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat Immunol 4, 741-8 (2003). Pubmed abstract # 12845325
- Caswell, P. T., Vadrevu, S. & Norman, J. C. Integrins: masters and slaves of endocytic transport. Nat Rev Mol Cell Biol 10, 843-53 (2009). Pubmed abstract # 19904298